Every patent application that unveils nucleotide and/or amino acid sequences must adhere to international filing standards by including sequence listings. The imminent implementation of ST.26, a new sequence listing standard effective from July 1, 2022, is anticipated to enhance the structural framework for seamless data exchange and validation among patent offices and biological sequence search databases. Despite ST.26 being an initiative by WIPO (World Intellectual Property Organization), the extent of accessibility of the new sequence listings on public portals rests with individual patent offices. The imperative for a new listing standard emerged due to the inadequacies of the current standard, ST.25, which needed more support for automated validation and data exchange.
In this article, we will delve into the disparities between ST.25 and ST.26, the drawbacks of ST.25, the modifications introduced by WIPO in the new standard, its advantages, and much more. However, before delving into these aspects, let us comprehensively understand what a sequence listing entails.
Understanding Sequence Listings: The Backbone of Biotech Patents
A sequence listing forms an integral part of patent applications, encompassing nucleotide and/or amino acid sequences. It stands as a prerequisite for patents within the biotech realm, covering innovations such as biomarkers, antibodies, oligonucleotides, and more. Adherence to prescribed filing regulations is essential to ensure smooth progress through the patent approval process. Presently, the prevailing standard for sequence listings is ST.25.
Now that we’ve clarified the concept of sequence listings, let’s delve into the associated rules applicable when submitting a patent application in the biotech sector.
Navigating Patent Rules: Your Roadmap to Sequence Listing Compliance
Below are key regulations regarding sequence listings in patent applications:
- Inventors are required to submit sequence listings in a computer-readable text format along with each patent application, disclosing either nucleotide or amino acid sequences.
- In cases where an international application reveals one or more nucleotide or amino acid sequences not provided in a computer-readable text format, the Searching Authority will issue a notice to the applicant for submission. Additionally, within one month, the applicant must pay the late furnishing fee as specified in the Fifth Schedule. Failure to comply with the notice will lead the Searching Authority to proceed with searching the international application without the sequence listing to a reasonable extent.
Need for a New Sequence Listing Standard
The necessity for implementing a new sequence listing standard arises from the deficiencies observed in the ST.25 practice. These inadequacies are outlined below:
- The ST.25 format failed to adhere to the requirements set forth by the International Nucleotide Sequence Database Collaboration (INSDC).
- Ambiguities existed in the rules, resulting in varying interpretations and enforcement by intellectual property offices worldwide.
- The ST.25 regulations lacked inclusion of sequence types th at have become common today, such as nucleotide analogs, D-amino acids, and branched sequences.
- Data organization within the ST.25 format was disorderly and unstructured.
- The ST.25 format presented challenges in terms of automated validation and facilitating data exchange.
Now, let’s delve into the alterations introduced by the new standard.
ST.26 Unpacked: What’s Different?
Key changes brought by WIPO through ST.26 are mentioned in Figure 1 below:
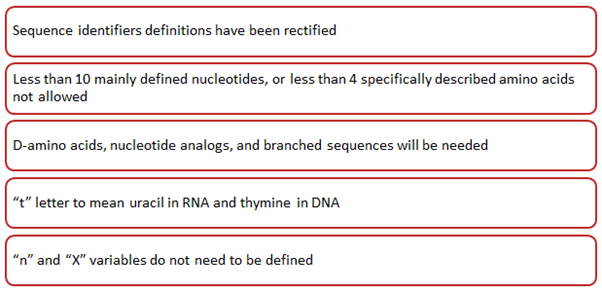
Additionally, WIPO, through ST.26, has implemented the following adjustments:
- Transition to .xml file types to address data loss concerns and facilitate data exchange readable by both humans and machines.
- Expanded sequence annotation options.
- Updated naming conventions and choices for organisms.
- Adopting three-letter codes instead of single-letter codes.
- Revised format for feature location.
- Prohibition of mixed-mode sequences.
- Inclusion of only the latest priority information, as opposed to all.
- Streamlined adjustments regarding language requisites.
ST.26 vs. ST.25: Spot the Difference
Let’s delve into how ST.26 has simplified the patent application process compared to its predecessor, ST.25. Here’s a breakdown of the differences:
| Contents | ST.25 Standard | ST.26 Standard |
| File Format | ASCII.txt with numeric identifiers | XML with elements and attributes |
| Permitted Sequence | Allows sequences: < 10 specific nucleotides < 4 specific amino acids | Prohibits sequences: < 10 specific nucleotides < 4 specific amino acids |
| Applicant/Inventor | All applicant and inventor names can be included | Only one applicant and optionally one innovator can be incorporated |
| Annotation | Annotation of sequences: Feature keys only | Annotation of sequences: Feature keys and qualifiers |
| D-amino acid/branched sequences/analogs | Not required to include D-amino acids, linear portions of branched sequences, nucleotide analogs | Must include D-amino acids, linear portions of branched sequences, nucleotide analogs |
| Priority application | All priority application info may be included | Only the earliest priority application info can be included |
| Title | One invention title permitted | Multiple invention titles permitted, each one in a different language |
| Applicant/inventor names | The name and invention titles of the applicant/inventor must be in basic Latin characters | The name of the applicant/inventor may be included using any valid Unicode character along with a basic Latin translation or transliteration |
| Type of Sequence | Sequences identified as RNA, DNA, or PRT only | Sequences identified as RNA, DNA, or AA along with a mandatory molecule type qualifier to further describe the molecule. For RNA: Genomic RNA, mRNA, RNA, rRNA, transcribed RNA. For DNA: Genomic DNA, unassigned DNA. For AA: Protein |
| Uracil Symbol | “u” represents uracil in nucleotide sequences | “t” represents thymine in DNA sequences and uracil in RNA sequences |
| Organism Name | Latin genus/species, Virus name “Artificial sequence”, “unknown” | Latin genus/species, Virus name “Synthetic construct”, “unidentified” |
| Variables | “n” and “Xaa” variables must have a definition provided in a feature | Default value assumed for “n” and “X” variables with no definition |
| AA Seq. Presentation | Amino acids sequences represented by three-letter abbreviations | Amino acids sequences represented by one-letter abbreviation |
| Mixed Mode Sequence | “Mixed mode” sequences permitted – nucleotide sequence with amino acid translation shown below | No “Mixed mode”; nucleotide translations are included in “translation” qualifiers only |
| Feature Location | Feature location format not clearly defined | Clearly defined feature location formats; permits use of “”, “^”, “join”, “order”, and “complement” in nucleotide sequences |
These distinctions shed light on how ST.26 has revamped the sequence listing standards, simplifying the patent application process for all stakeholders involved.
The Benefits of Upgrading: Why ST.26 Rocks
Several benefits of using the ST.26 standard for your sequence listings are listed below:
- Embracing the new standard enables applicants to consolidate multiple sequence listings into a single patent application, whether for international, national, or regional procedures.
- ST.26 elevates the precision and caliber of sequence presentations, ensuring greater accuracy in conveying genetic information.
- Facilitating seamless data searches, ST.26 simplifies the process of accessing sequence data, enhancing efficiency in research and patent examination.
- With ST.26, sequence data can be effortlessly exchanged electronically and integrated into digital databases, promoting accessibility and collaboration in the scientific community.
- ST.26 fosters harmonization among intellectual property offices, ensuring consistency and coherence in sequence listing standards globally.
- By enhancing automation in data validation and streamlining processing, ST.26 accelerates the patent approval process, reducing administrative burdens and expediting innovation.
- Leveraging the structured format of XML sequence listings, ST.26 enhances the quality of submissions, minimizing errors and improving overall presentation standards.
Prepare for ST.26 Success – A Guide to Compliant Sequence Listings!
You can utilize the WIPO Sequence tool to transform outdated ST.25 TXT sequence listings into the latest ST.26 XML format. It’s the responsibility of the applicant to share sequence listings according to the new standard, as the Patent Office does not handle the conversion. Additionally, WIPO Sequence can be utilized to identify errors in the ST.26 sequence and view the new standard’s XML file in a human-readable format directly in the browser.
WIPO Sequence: Your Gateway to ST.26 Compliance!
WIPO Sequence stands as a global software solution empowering patent applicants to craft amino acid and nucleotide sequence listings that adhere to the WIPO ST.26 standard, whether for national or international patent applications. Developed by WIPO, this tool facilitates the authoring, validation, and generation of ST.26 compliant sequence listings. Below are some key features of WIPO Sequence:
- It eradicates the necessity of editing XML files.
- Users can securely store sequence information within a project, validate it, and generate a sequence listing in the ST.26-compliant format.
- It offers seamless import capabilities for data from various sources, including ST.26 sequence listings, projects, ST.25 sequence listings, multi-sequence format files, raw format files, and FASTA format files.
- The tool enables the validation of sequence listings in XML format.
- It provides support for the export and import of XLIFF files, commonly used by translators.
Conclusion: Embracing ST.26 for a Brighter Patent Future!
In the biotechnology sector, patent applications require sequence listings adhering to WIPO’s standardized format. Previously, applicants followed the ST25 standard, but as of July 1, 2022, the new ST26 standard has been implemented. The transition from ST25 to ST26 aims to enhance accuracy and quality in presentations, streamline sequence data searches, and facilitate the electronic exchange of sequence data.
Non-compliance with WIPO’s sequence listing standards may lead to the rejection of a patent application. The Sequence Listing Company’s team of patent experts stands ready to assist you in preparing compliant sequence listings. Explore our sequence listing service page for further information.
